This December, members of the Hehnly Lab traveled to Philadelphia to present their work at the 2025 American Society for Cell Biology (ASCB) Annual Meeting.
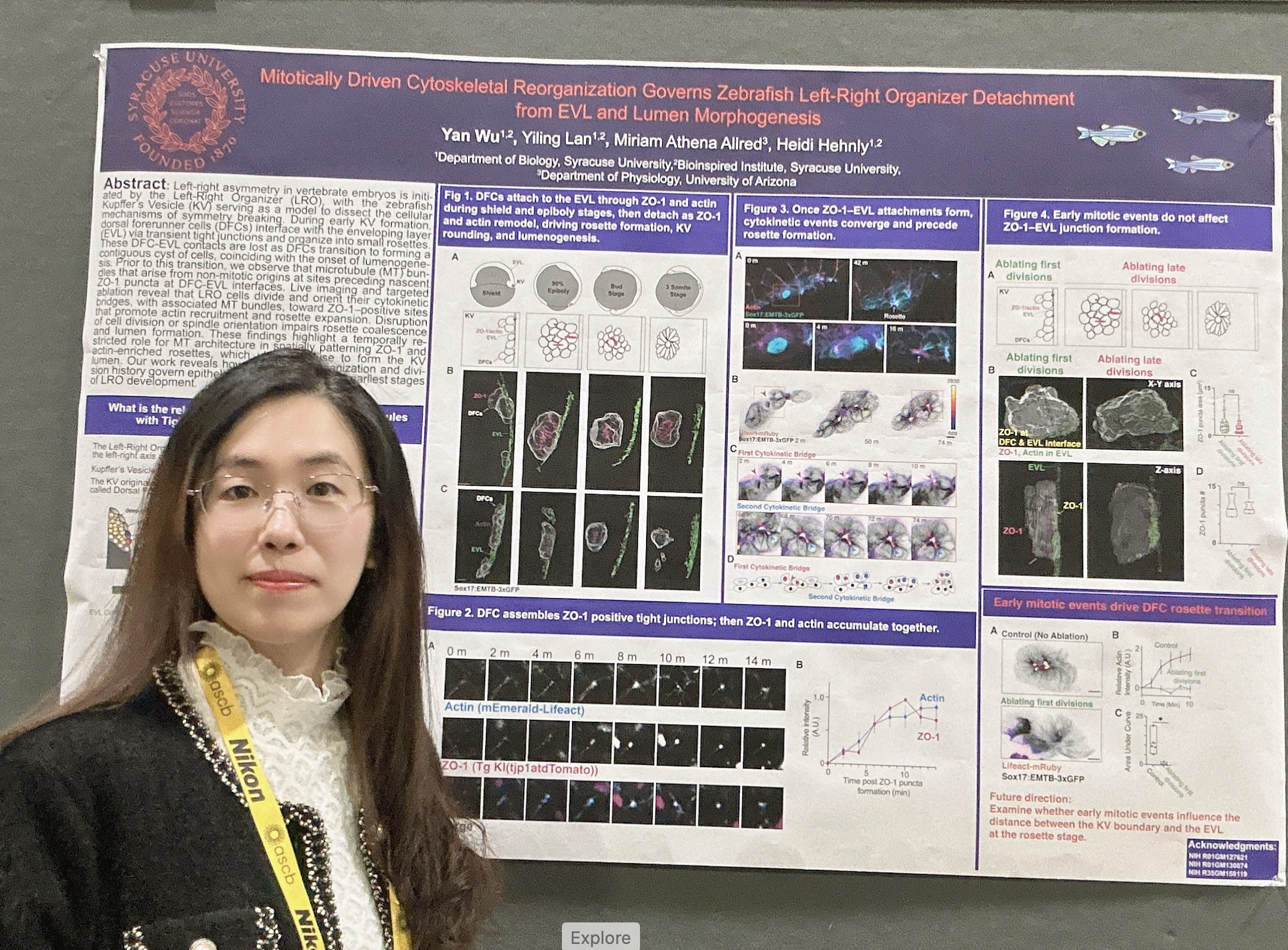
Yan Wu presented her poster, “Mitotically driven cytoskeletal reorganization governs zebrafish left–right organizer detachment from the EVL and lumen morphogenesis.” This work highlights how dynamic mitotic events shape tissue architecture during early embryogenesis and reflects a major collaborative effort within the lab. In addition to Yan’s leadership on the project, Yiling Lan and summer undergraduate Miriam Athena Allred made substantial experimental and conceptual contributions, with additional contributions from Carys Timpson during her summer research period.
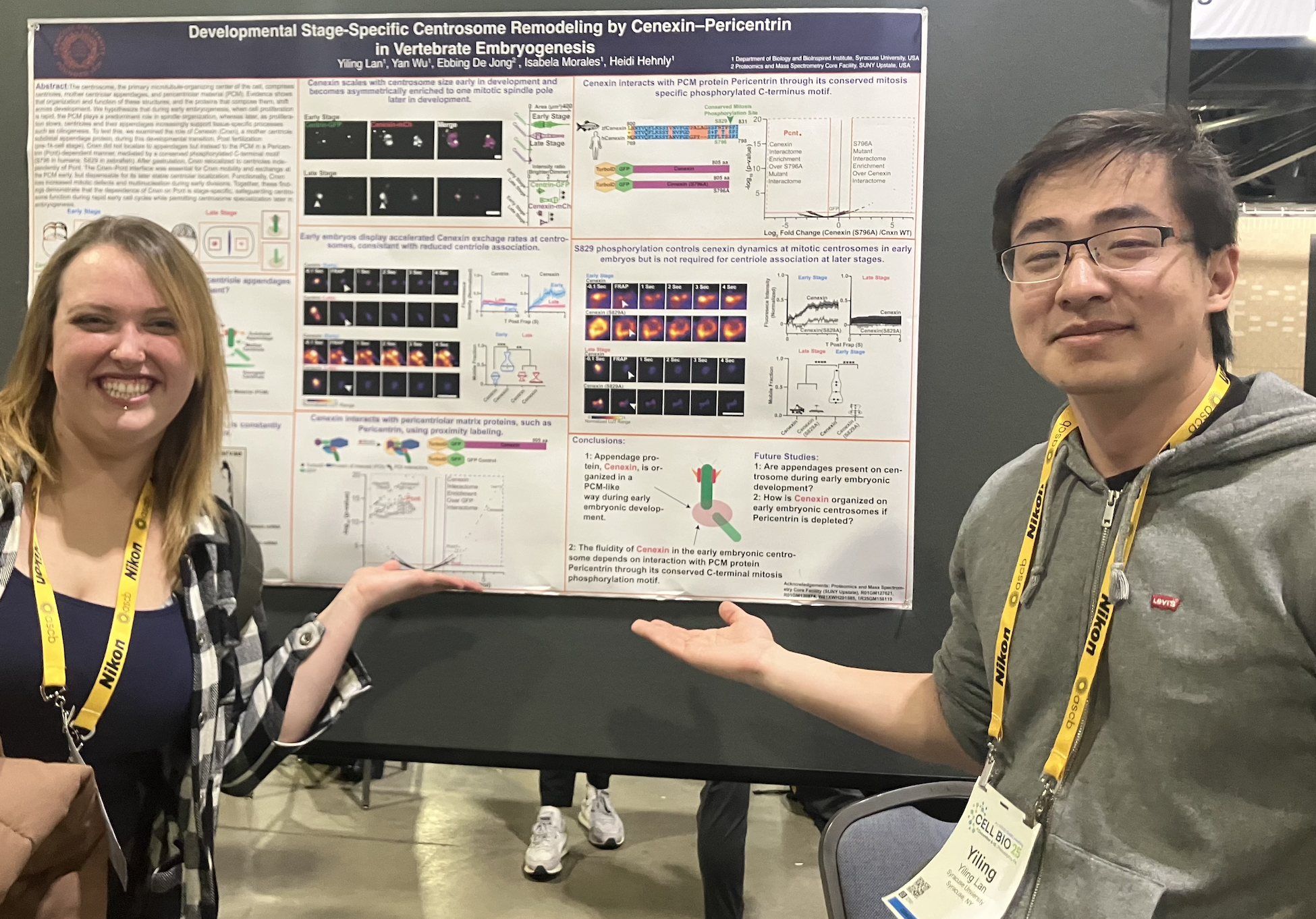
Yiling Lan also presented his work, “Developmental stage–specific centrosome remodeling by cenexin–pericentrin in vertebrate embryogenesis,” which explores how centrosome composition and organization are developmentally regulated to support morphogenesis. While at ASCB, Yiling had the opportunity to reconnect with former lab member Erin Curtis, now completing her PhD jointly at MIT and Duke— a reminder of the extended scientific community that grows from shared training experiences.
We were also excited to have Albert Adhya attend ASCB as he begins his journey in the lab. For Albert, the meeting served as an immersive introduction to the broader cell biology community and an opportunity to see firsthand how scientific ideas are communicated and refined. The group capped off the meeting with a celebratory dinner at Morimoto—an excellent way to mark a successful and energizing conference.